Research
Gene Regulation during seed development
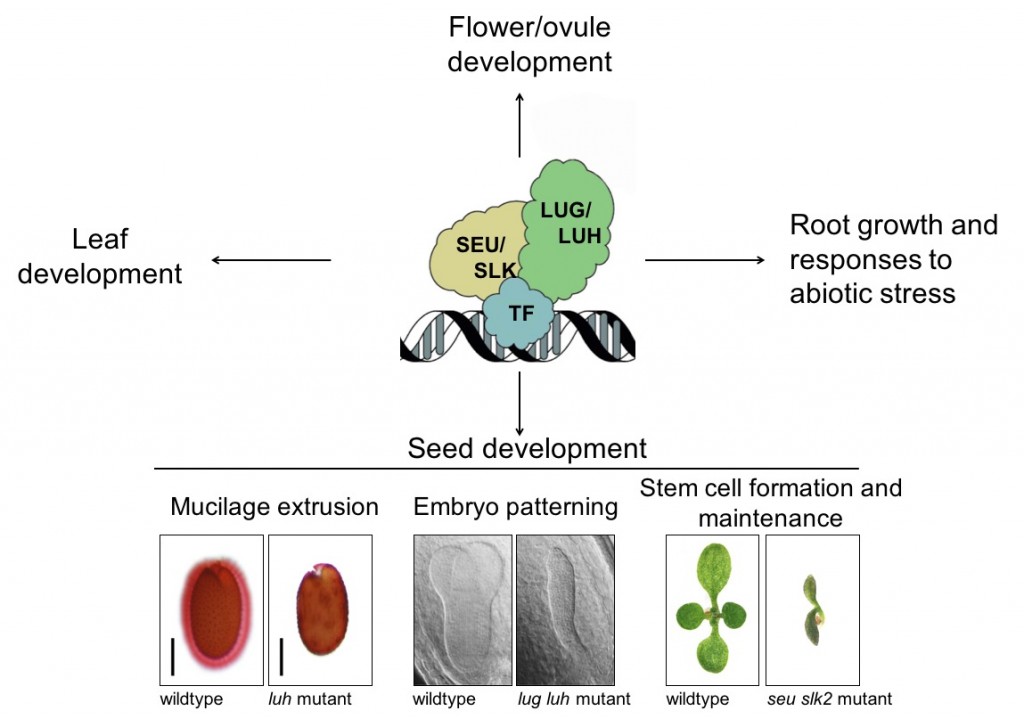
We have shown that the Gro/TUP1-like co-repressors LEUNIG (LUG) and LEUNIG_HOMOLOG (LUH) control diverse processes in the plant including apical-basal and auxin patterning during early embryogenesis, cell fate acquisition in developing leaves, and pectin modification in the developing seed coat (Stahle et al., 2009; Walker et al., 2011; Lee et al., 2014). As LUG/LUH lack a DNA-binding domain, they must interact with DNA binding co-factors if they are to be recruited to the regulatory sequences of target genes. This may occur through direct interactions with transcription factors or indirectly via the adaptor proteins SEUSS (SEU) or SEU-LIKE proteins (SLKs). As a result of these interactions, LUG/LUH are part of a large regulatory complex.
Current research projects
- Using RNA-seq and chromatin immunoprecipitation to identify the network of genes controlled by the LUG/LUH regulatory complex during embryonic and post-embryonic development
- Using protein interaction assays to define the interactome of LUG/LUH and associated co-regulators SEU/SLKs
Biotechnology – Seed size control
Seed size is a critical trait that affects the long-term survival of plants in the wild and yield potential of our most important crops. Seeds are a unique combination of maternal tissues (the seed coat) with filial tissues derived from double fertilization of the ovules by pollen (the diploid embryo and triploid endosperm). Thus, seed development is a fascinating and complex process that requires coordination between tissues of different origin and ploidy.
Recent studies have shown that the seed coat plays an important role in regulating seed size with various models being proposed to explain how this is achieved. Our studies of gene regulation in the developing Arabidopsis seed coat have uncovered a new regulatory pathway involved in the control of seed size. We are now developing the tools that will allow us to explore this phenotype in greater detail.
Current research projects
- Developing molecular tools to alter gene activity in specific layers of the seed coat so that relative contribution of each seed coat layer to the regulation of seed size can be assessed.
- Using knowledge of seed size regulation in Arabidopsis to alter seed size and yield in crop plants such as Brassica napus (canola).
Biotechnology – Transformation technology
Although crop improvements have traditionally been achieved through selective breeding, the advent of gene-editing technologies means it is now possible to generate genetic variation in a much more targeted manner, in the germplasm of choice and in a shorter timeframe. Unfortunately this approach is not possible for plant species where genetic transformation approaches are either not possible (such as the superfood chia) or where they rely on passage through tissue culture which is both time consuming, expensive and prone to generate undesirable genetic variation.
Current research projects
- Developing transformation protocols for recalcitrant species such as Chia so that gene-editing technology can be deployed in this species (a collaborative effort with the Roessner and Ebert groups within the School of BioSciences)
- Developing a faster and lest costly genetic transformation approach for Canola, which will resolve bottleneck issues associated with gene-editing in this crop species.