Fresh publications!
Climate contributes to the evolution of pesticide resistance | James Maino, Paul Umina, Ary Hoffmann
Abstract
The evolution of pesticide resistance through space and time is of great economic significance to modern agricultural production systems, and consequently, is often well documented. It can thus be used to dissect the evolutionary and ecological processes that underpin large-scale evolutionary responses. There are now nearly 600 documented cases of pesticide resistance in arthropod pests. Although the evolution of resistance is often attributed to the persistent use of chemicals for pest suppression, the rate of development of resistance should also depend on other factors, including climatic conditions that influence population size and generation time. Here, we test whether climatic variables are linked to evolution of resistance by examining the spatial pattern of pyrethroid resistance in an important agricultural pest.
Can high-throughput sequencing detect macroinvertebrate diversity for routine monitoring of an urban river? | Melissa Carew, Claudette Kellar, Vincent Pettigrove, Ary Hoffmann
Abstract
Macroinvertebrate families identified through morphological examination have traditionally been used in routine assessment of freshwater ecosystems. However, high throughput DNA sequencing (HTS) promises to improve routine assessment by providing rapid and cost-effective identification of macroinvertebrate species. In freshwater ecosystems in urbanised areas where family diversity is often low, new insights into ecosystem condition and impacting factors are likely through species-level assessments. Here we compare morphological identification to HTS based identification of macroinvertebrate families by considering 12 sites in an urban river system. Some taxa detected morphologically were not detected by HTS and vice versa. However, this had only a small impact on computed family-level metrics of ecological condition. We detected multiple species using HTS in the Chironomidae, Coenagrionidae, Hydrobiidae, Leptoceridae, Ceratopogonidae, Corixidae, Veliidae, Oligochaeta and Acarina. The highest species diversity was found in the Chironomidae, and for many of these species we had prior knowledge of their likely pollution sensitivity. In the Chironomidae, we showed that species level data was congruent with expectations based on measured levels of pollutants at sites and other family level metrics. Importantly, we also identified many species in the same family that differed in their distribution and likely pollution sensitivity in this urban river system. Therefore, HTS provided similar levels of information to traditional methods at the family level, but also generated new information for more accurate condition monitoring at the species level.
Laboratory selection for resistance to sulfoxaflor and fitness costs in the green peach aphid Myzus persicae | Ze-Hua Wang, Ya-Jun Gong, Jin-Cui Chen, Xiao-Chun Su, Li-Jun Cao, Ary Hoffmann, Shu-Jun Wei
Abstract
Sulfoxaflor is a newly released fourth-generation neonicotinoid insecticide for management of sap-feeding pests that have developed resistance to established insecticide groups. The risk of resistance developing to this pesticide in target pests is unclear. We selected a strain of the green peach aphid, Myzus persicae (Sulzer) (Hemiptera: Aphididae), for resistance to sulfoxaflor in the laboratory, which showed 199-fold resistance after 45 generations compared to the starting population. Life table analysis showed that the resistant strain had a fitness of 0.83 compared to the susceptible strain. Adult longevity of the resistant strain was reduced by 9.55% compared to the susceptible strain. The period when adults of the resistant strain produced offspring was reduced by 17.19%, while the mean fecundity of the resistant strain was reduced by 15%. These findings suggest that M. persicae can develop a high level of resistance to sulfoxaflor, but fitness costs may result in a recovery of sensitivity when field populations are no longer exposed to sulfoxaflor.
Influence of previous host plants on the reproductive success of a polyphagous mite pest, Halotydeus destructor (Trombidiformes: Penthaleidae) | Xuan Cheng, Paul Umina, Ary Hoffmann, Dominic Reisig
Abstract
In the evolution of phytophagous arthropods, adaptation to a single type of host plant is generally assumed to lead to a reduction in fitness on other host plant types, resulting in increasing host specialization. While this process is normally considered to be genetically based, short-term effects acting within one generation (plasticity) or across two generations (cross-generation variation) could also play a role. Here, we test these effects in the redlegged earth mite, Halotydeus destructor (Tucker) (Prostigmata: Penthaleidae), a major agricultural pest of multiple crop plants. Field populations of mites were collected from grasses, legumes, and broad-leaf weeds and placed into enclosures with different plant types. The survival, net reproductive output (Ro), and feeding damage of each mite population were assessed across two generations. The interaction between the origin of mites and plant type had a significant effect on parental survival, Ro, offspring development, and feeding damage. Mites collected from legumes showed higher parental survival on all host types; however, Ro, offspring development and feeding damage were all higher when mites were placed onto the same plant type from which they were collected. These patterns point to the ability of H. destructor to perform well on host plants even in the absence of genetically differentiated host races, but also the likelihood of performance trade-offs when populations are forced to rapidly change hosts within and across sequential generations.
The Wolbachia strain wAu provides highly efficient virus transmission blocking in Aedes aegypti | Thomas Ant, Christie Herd, Vincent Geoghegan, Ary Hoffmann, Steven Sinkins
Abstract
Introduced transinfections of the inherited bacteria Wolbachia can inhibit transmission of viruses by Aedes mosquitoes, and in Ae. aegypti are now being deployed for dengue control in a number of countries. Only three Wolbachia strains from the large number that exist in nature have to date been introduced and characterized in this species. Here novel Ae. aegypti transinfections were generated using the wAlbA and wAu strains. In its native Ae. albopictus, wAlbA is maintained at lower density than the co-infecting wAlbB, but following transfer to Ae. aegypti the relative strain density was reversed, illustrating the strain-specific nature of Wolbachia-host co-adaptation in determining density. The wAu strain also reached high densities in Ae. aegypti, and provided highly efficient transmission blocking of dengue and Zika viruses. Both wAu and wAlbA were less susceptible than wMel to density reduction/incomplete maternal transmission resulting from elevated larval rearing temperatures. Although wAu does not induce cytoplasmic incompatibility (CI), it was stably combined with a CI-inducing strain as a superinfection, and this would facilitate its spread into wild populations. Wolbachia wAu provides a very promising new option for arbovirus control, particularly for deployment in hot tropical climates.
Fine-scale landscape genomics helps explain the slow spatial spread of Wolbachia through the Aedes aegypti population in Cairns, Australia | Thomas Schmidt, Igor Filipović, Ary Hoffmann, Gordana Rašic
Abstract
The endosymbiotic bacterium Wolbachia suppresses the capacity for arbovirus transmission in the mosquito Aedes aegypti, and can spread spatially through wild mosquito populations following local introductions. Recent introductions in Cairns, Australia have demonstrated slower than expected spatial spread. Potential reasons for this include: (i) barriers to Ae. aegypti dispersal; (ii) higher incidence of long-range dispersal; and (iii) intergenerational loss of Wolbachia. We investigated these three potential factors using genome-wide single-nucleotide polymorphisms (SNPs) and an assay for the Wolbachia infection wMel in 161 Ae. aegypti collected from Cairns in 2015. We detected a small but significant barrier effect of Cairns highways on Ae. aegypti dispersal using distance-based redundancy analysis and patch-based simulation analysis. We detected a pair of putative full-siblings in ovitraps 1312 m apart, indicating long-distance female movement likely mediated by human transport. We also found a pair of full-siblings of different infection status, indicating intergenerational loss of Wolbachia in the field. These three factors are all expected to contribute to the slow spread of Wolbachia through Ae. aegypti populations, though from our results it is unclear whether Wolbachia loss and long-distance movement are sufficiently common to reduce the speed of spatial spread appreciably. Our findings inform the strategic deployment of Wolbachia-infected mosquitoes during releases, and show how parameter estimates from laboratory studies may differ from those estimated using field data. Our landscape genomics approach can be extended to other host/symbiont systems that are being considered for biocontrol.
Mission Accomplished? We Need a Guide to the ‘Post Release’World of Wolbachia for Aedes-borne Disease Control | Scott Ritchie, Andrew van den Hurk, Michael Smout, Kyran Staunton, Ary Hoffmann
Abstract
Historically, sustained control of Aedes aegypti , the vector of dengue, chikungunya, yellow fever, and Zika viruses, has been largely ineffective. Subsequently, two novel ‘rear and release’ control strategies utilizing mosquitoes infected with Wolbachia are currently being developed and deployed widely. In the incompatible insect technique, male Aedes mosquitoes, infected with Wolbachia , suppress populations through unproductive mating. In the transinfection strategy, both male and female Wolbachia -infected Ae. aegypti mosquitoes rapidly infect the wild population with Wolbachia , blocking virus transmission. It is critical to monitor the long-term stability of Wolbachia in host populations, and also the ability of this bacterium to continually inhibit virus transmission. Ongoing release and monitoring programs must be future-proofed should political support weaken when these vectors are successfully controlled
Incidence of facultative bacterial endosymbionts in spider mites associated with local environment and host plant | Yu-Xi Zhu, Yue-Ling Song, Yan-Kai Zhang, Ary Hoffmann, Jin-Cheng Zhou, Jing-Tao Sun, Xiao-Yue Hong
Abstract
Spider mites are frequently associated with multiple endosymbionts whose infection patterns often exhibit spatial and temporal variation. However, the association between endosymbiont prevalence and environmental factors remains unclear. Here, we surveyed endosymbionts in natural populations of the spider mite, Tetranychus truncatus, in China, screening 935 spider mites from 21 localities and 12 host plant species. Three facultative endosymbiont lineages, Wolbachia, Cardinium, and Spiroplasma, were detected at different infection frequencies (52.5%, 26.3%, and 8.6%, respectively). Multiple endosymbiont infections were observed in most local populations, and the incidence of individuals with the Wolbachia-Spiroplasma co-infection was higher than expected from the frequency of each infection within a population. Endosymbiont infection frequencies exhibited associations with environmental factors: Wolbachia infection rates increased at localities with higher annual mean temperature, while Cardinium and Spiroplasma infection rates increased at localities from higher altitudes. Wolbachia was more common in mites from Lycopersicon esculentum and Glycine max compared to Zea mays. This study highlights that host-endosymbiont interactions may be associated with environmental factors, including climate and other geographically linked factors, as well as the host’s food plant.
Life‐history traits and physiological limits of the alpine fly Drosophila nigrosparsa (Diptera: Drosophilidae): A comparative study | Martin‐Carl Kinzner, Patrick Krapf, Martina Nindl, Carina Heussler, Stephanie Eisenkölbl, Ary Hoffmann, Julia Seeber, Wolfgang Arthofer, Birgit Schlick‐Steiner, Florian Steiner
Abstract
Interspecific variation in life-history traits and physiological limits can be linked to the environmental conditions species experience, including climatic conditions. As alpine environments are particularly vulnerable under climate change, we focus on the montane-alpine fly Drosophila nigrosparsa. Here, we characterized some of its life-history traits and physiological limits and compared these with those of other drosophilids, namely Drosophila hydei, Drosophila melanogaster, and Drosophila obscura. We assayed oviposition rate, longevity, productivity, development time, larval competitiveness, starvation resistance, and heat and cold tolerance. Compared with the other species assayed, D. nigrosparsa is less fecund, relatively long-living, starvation susceptible, cold adapted, and surprisingly well heat adapted. These life-history characteristics provide insights into invertebrate adaptations to alpine conditions which may evolve under ongoing climate change.
Environmental variation partitioned into separate heritable components | Michael Ørsted, Palle Duun Rohde, Ary Hoffmann, Peter Sørensen, Torsten Nygaard Kristensen
Abstract
Trait variation is normally separated into genetic and environmental components, yet genetic factors also control the expression of environmental variation, encompassing plasticity across environmental gradients and within-environment responses. We defined four components of environmental variation: plasticity across environments, variability in plasticity, variation within environments, and differences in within-environment variation across environments. We assessed these components for cold tolerance across five rearing temperatures using the Drosophila melanogaster Genetic Reference Panel (DGRP). The four components were found to be heritable, and genetically correlated to different extents. By whole genome single marker regression, we detected multiple candidate genes controlling the four components and showed limited overlap in genes affecting them. Using the binary UAS-GAL4 system, we functionally validated the effects of a subset of candidate genes affecting each of the four components of environmental variation and also confirmed the genetic and phenotypic correlations obtained from the DGRP in distinct genetic backgrounds. We delineate selection targets associated with environmental variation and the constraints acting upon them, providing a framework for evolutionary and applied studies on environmental sensitivity. Based on our results we suggest that the traditional quantitative genetic view of environmental variation and genotype-by-environment interactions needs revisiting.
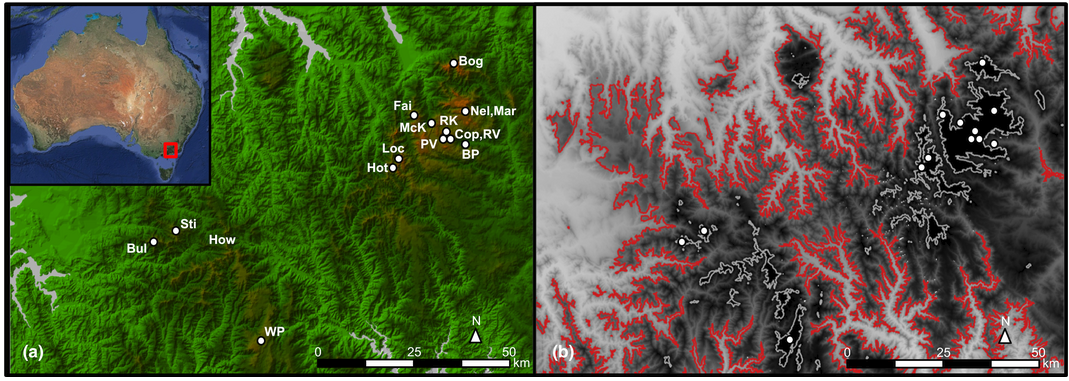
Spatial patterns of genetic diversity among Australian alpine flora communities revealed by comparative phylogenomics | Nick Bell, Philippa Griffin, Ary Hoffmann, Adam Miller
Abstract
The alpine region of mainland Australia is one of the world’s 187 biodiversity hotspots. Genetic analyses of Australian alpine fauna indicate high levels of endemism on fine spatial scales, unlike Northern Hemisphere alpine systems where shallow genetic differentiation is typically observed among populations. These discrepancies have been attributed to differences in elevation and influence from glacial activity, and point to a unique phylogeographic history affecting Australian alpine biodiversity. To test generality of these findings across Australian alpine biota, we assessed patterns of genetic structure across plant species.
There is deep lineage diversification between the North and Central Victorian Alpine regions, reflecting a high level of endemism. These findings differ from those reported for alpine biodiversity from New South Wales and much of the Northern Hemisphere, and support the notion that genetic diversity is typically greatest in areas least affected by historical ice sheet formation. We discuss the implications of our findings in the context of conservation planning, and highlight the benefits of this rapid and cost-effective genome scan approach for characterizing evolutionary processes at multispecies and landscape scales.