Using genomics to determine the adaptive potential of populations
Words: Ary Hoffmann
One of the central tenets of conservation biology is that high levels of genetic variation in natural populations is important for their long-term survival. High levels of variation allow populations to adapt to changing environments through evolution. This process is now recognised as being extremely fast when selection pressures are high. In fact many populations of animals and plants are undergoing a continuous process of genetic adaptation to many different types of environmental variables. The genetic changes in adaptation typically involve the cumulative effects of many genes, each with small effects on the traits being selected.
While the importance of this process is well recognised, assessing the evolutionary genetic potential of populations is challenging. One common approach is to measure levels of past and current inbreeding in populations, on the understanding that inbreeding causes exposure of deleterious recessive mutations, thereby decreasing fitness. In addition, the small size of populations required to produce inbreeding effects is expected to lead to the loss of adaptive alleles through a random process of allele sorting known as genetic drift.
Another possible approach to measure evolutionary adaptation potential is to evaluate genetic variation across the entire genome using many molecular markers. By characterising thousands of markers, it is possible to obtain a representative picture of genetic variation across the entire genome. This in turn is expected to be linked to adaptive evolutionary potential, given that many genes across the genome will be involved in adaptive processes, and even though the markers surveyed might not necessarily themselves be connected to adaptation.
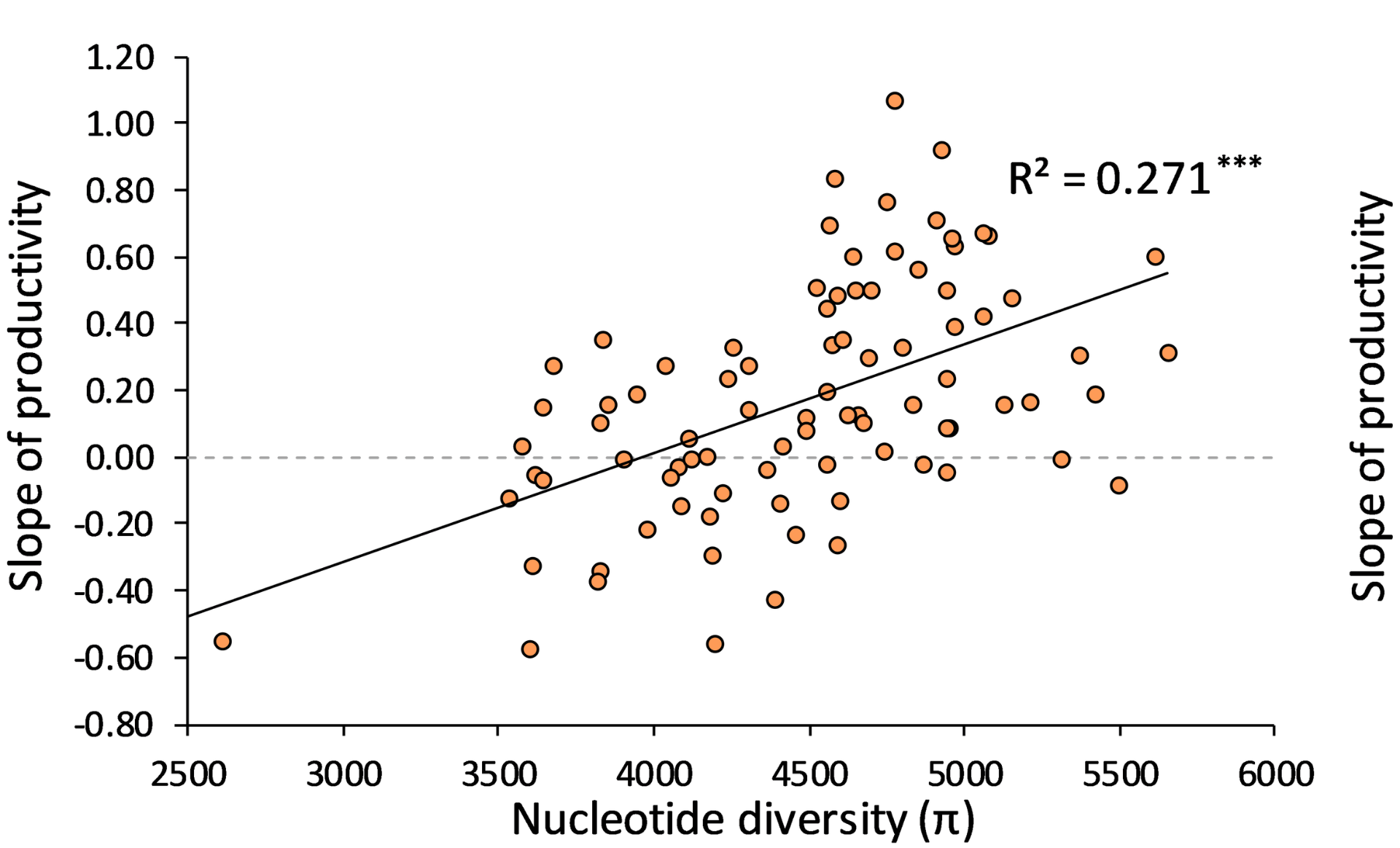
To test these ideas, work initiated in our lab and then completed in Aalborg Denmark undertook a detailed assessment of adaptation rates and genomic variation in a set of Drosophila lines that were exposed to stressful feeding conditions involving low nutrition and low pH. We measured genomic variation by examining 11000-36000 SNPs across the genome and exposed the fly lines to increasing levels of stress. The lines gradually adapted, increasing their fitness under stressful conditions.
The rates of adaptation in these lines varied massively. We then tested which features of the populations best predicted their rates of adaptation, the history of prior inbreeding to which the populations had been exposed or the genomic variation present in the populations. The results provided very clear support for the latter (Figure). Lines with high levels of genomic variation responded much better to selection than lines with low levels of genomic variation.
The strong association between genomic variation (nucleotide diversity) and rate of adaptation (slope showing increase in productivity achieved as flies adapted to stressful medium)
These findings not only reinforce the importance of genetic variation in populations when responding to an environmental change, but also point to a simple way of measuring this potential given that SNP screens are now relatively cheap to complete in a population. When deciding on the conservation value of populations, our work suggests that levels of genomic variation should be screened. This in turn indicates whether the conserved populations are likely to respond to future stresses through evolution, and thereby avoid extinction.
This work has now been published open access in PLoS Genetics