
Using unsorted sweep-net samples to rapidly assess macroinvertebrate biodiversity
Words: Melissa Carew
Freshwater invertebrates are the insects, snails, clams, mites, crustaceans, and worms that inhabit streams, rivers, ponds and wetlands. They play an important role in understanding the health of our freshwater environments. The biodiversity of invertebrates present in freshwaters is routinely used by water managers to assess the ecological condition and feeds into decision making about stream management.
Biodiversity of freshwater invertebrates is traditionally determined by identifying invertebrates using taxonomic keys to the family level of taxonomic classification. Finer levels of taxonomic identification, such as identifying invertebrates to species, is not widely used as it is difficult, even impossible to identify some families using physical differences to distinguish species. However, using invertebrate DNA we can achieve finer levels of identification routinely and cost-effectively. Applying a process known as ‘DNA metabarcoding’ to bulk samples of freshwater invertebrates allows them to be rapidly identify to species. This provides an understanding of species distributions and how their biodiversity changes along environmental gradients and in response to human-induced changes.
Developing standardised, robust and cost-effective protocols that use DNA metabarcoding is important for water managers to routinely use DNA technologies. Traditionally, freshwater invertebrates are collected in nets and sorted out from sampling debris, which can take considerable time. However, we tested the use of DNA metabarcoding on unsorted invertebrates (invertebrates and debris) from net samples collected from stream pool or edge habitats in order to further reduce the cost and time it taken to process samples. We processed these unsorted samples with a standardised method and tested different DNA amplification primer sets to target the DNA from invertebrates for DNA metabarcoding.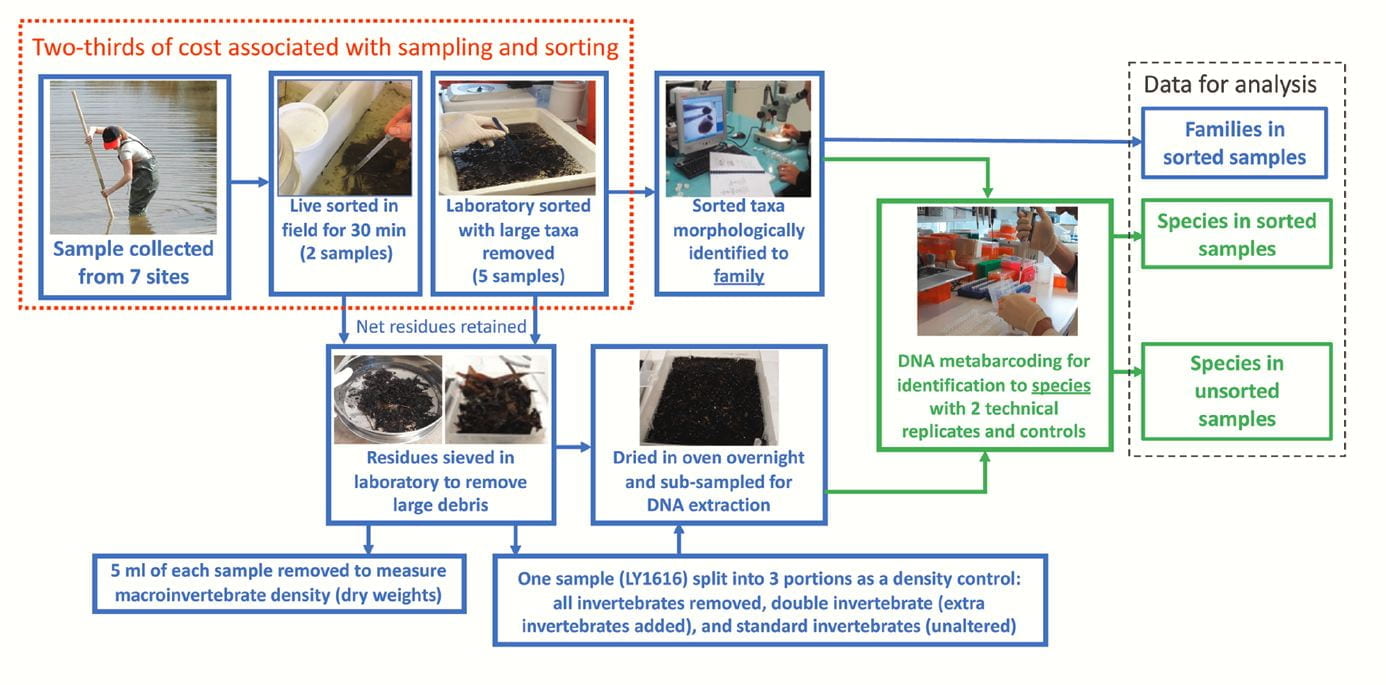
Workflow for collecting and processing samples used in this evaluation. Blue boxes refer to the work plan and sampling design, whereas green boxes refer to the DNA metabarcoding component. Sampling and sorting components that cover ⅔ of total costs (taken from Elbrecht et al. 2017) are in the red dotted box.
We found that currently available invertebrate DNA amplification primers were well suited for processing unsorted samples, though some performed better than others. However, invertebrate density (the ratio of invertebrates to net debris) affected the number of species detected with DNA metabarcoding, particularly the detection of rare taxa. Our study has shown that DNA metabarcoding of unsorted net contents could streamline macroinvertebrate sample processing for water managers providing a new and innovative way to understand and monitor the biodiversity of invertebrates in our freshwater environments.
This research was published online July 29, 2021, in Freshwater Science https://doi.org/10.1086/716215